![PDF] Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences by Weizhong Li, Adam Godzik · 10.1093/bioinformatics/btl158 · OA.mg PDF] Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences by Weizhong Li, Adam Godzik · 10.1093/bioinformatics/btl158 · OA.mg](https://og.oa.mg/Cd-hit%3A%20a%20fast%20program%20for%20clustering%20and%20comparing%20large%20sets%20of%20protein%20or%20nucleotide%20sequences.png?author=%20Weizhong%20Li,%20Adam%20Godzik)
PDF] Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences by Weizhong Li, Adam Godzik · 10.1093/bioinformatics/btl158 · OA.mg
De Novo Assembly of the Transcriptome of the Non-Model Plant Streptocarpus rexii Employing a Novel Heuristic to Recover Locus-Specific Transcript Clusters | PLOS ONE

kClust: fast and sensitive clustering of large protein sequence databases | BMC Bioinformatics | Full Text

Fast Program for Clustering and Comparing Large Sets of Protein or Nucleotide Sequences | SpringerLink

An example illustrating how the CD-HIT main paradigm works. Record 1 is... | Download Scientific Diagram
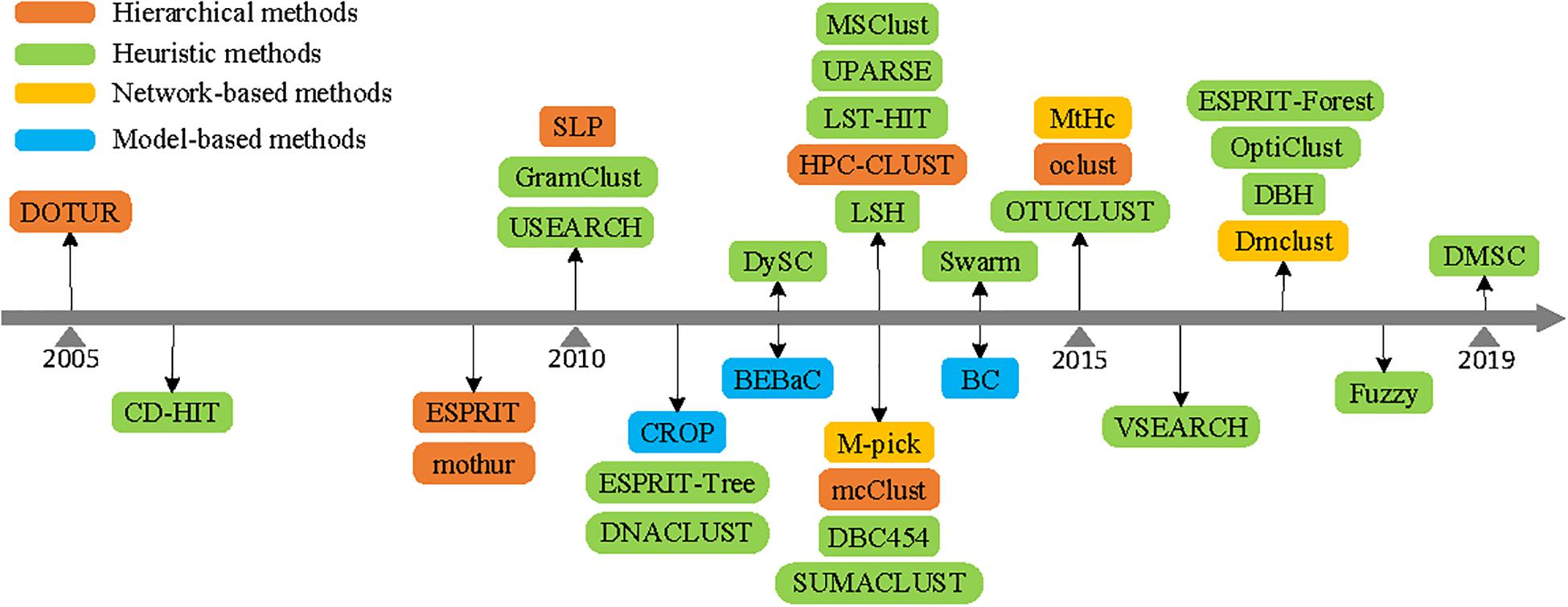
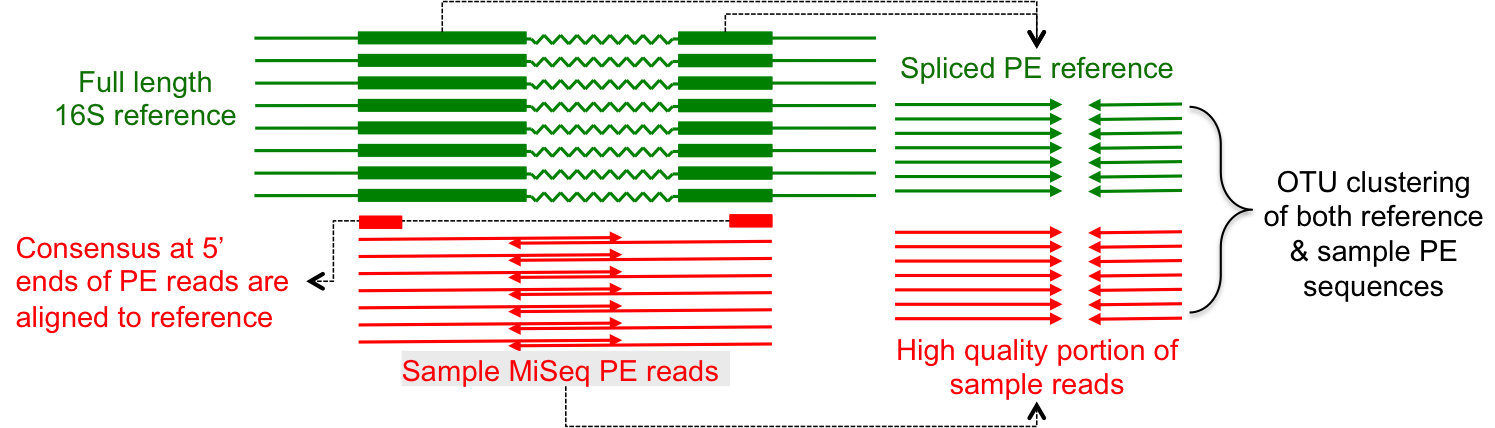
Frontiers | Comparison of Methods for Picking the Operational Taxonomic Units From Amplicon Sequences

An example illustrating how the CD-HIT main paradigm works. Record 1 is... | Download Scientific Diagram

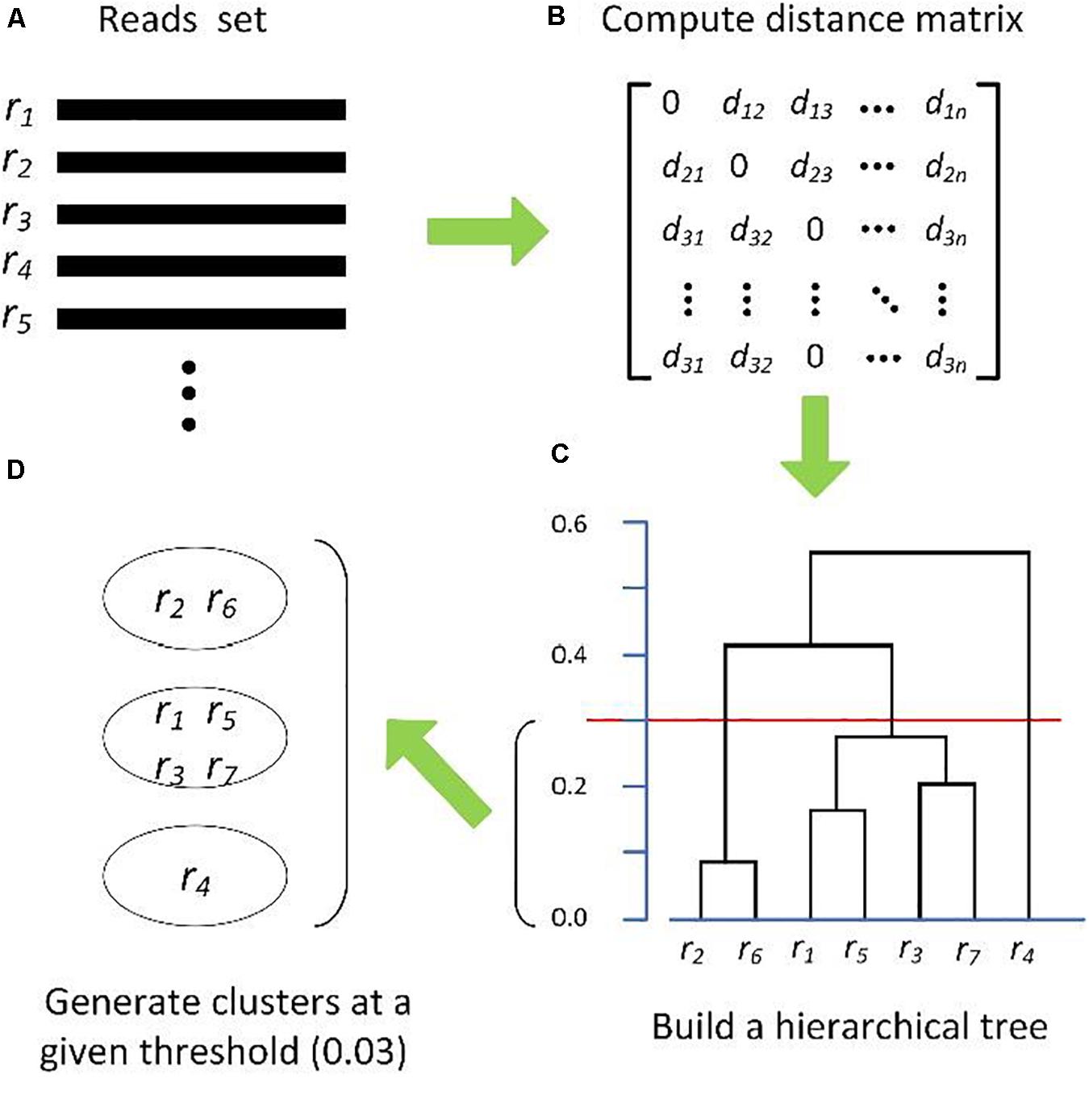
Clustering biological sequences with dynamic sequence similarity threshold | BMC Bioinformatics | Full Text
![PDF] Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences | Semantic Scholar PDF] Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/f77ee5140d2c9408165c6cb756344fa6b0067f10/2-Table1-1.png)
PDF] Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences | Semantic Scholar
Frontiers | Comparison of Methods for Picking the Operational Taxonomic Units From Amplicon Sequences
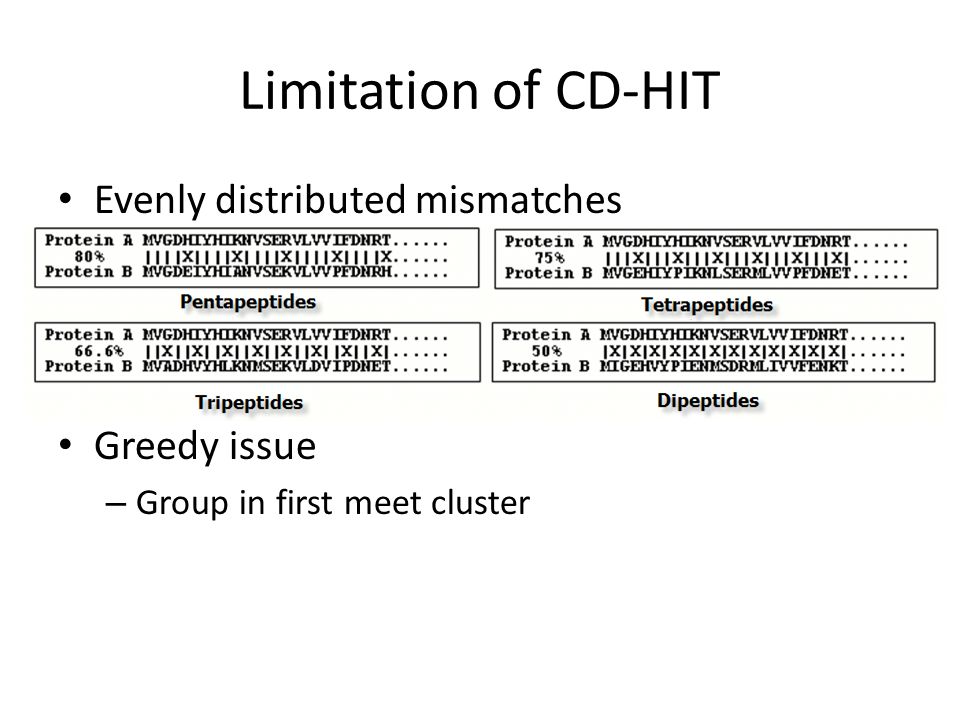
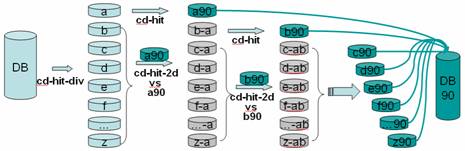
Analysis and comparison of very large metagenomes with fast clustering and functional annotation Weizhong Li, BMC Bioinformatics 2009 Present by Chuan-Yih. - ppt download

Fast Program for Clustering and Comparing Large Sets of Protein or Nucleotide Sequences | SpringerLink
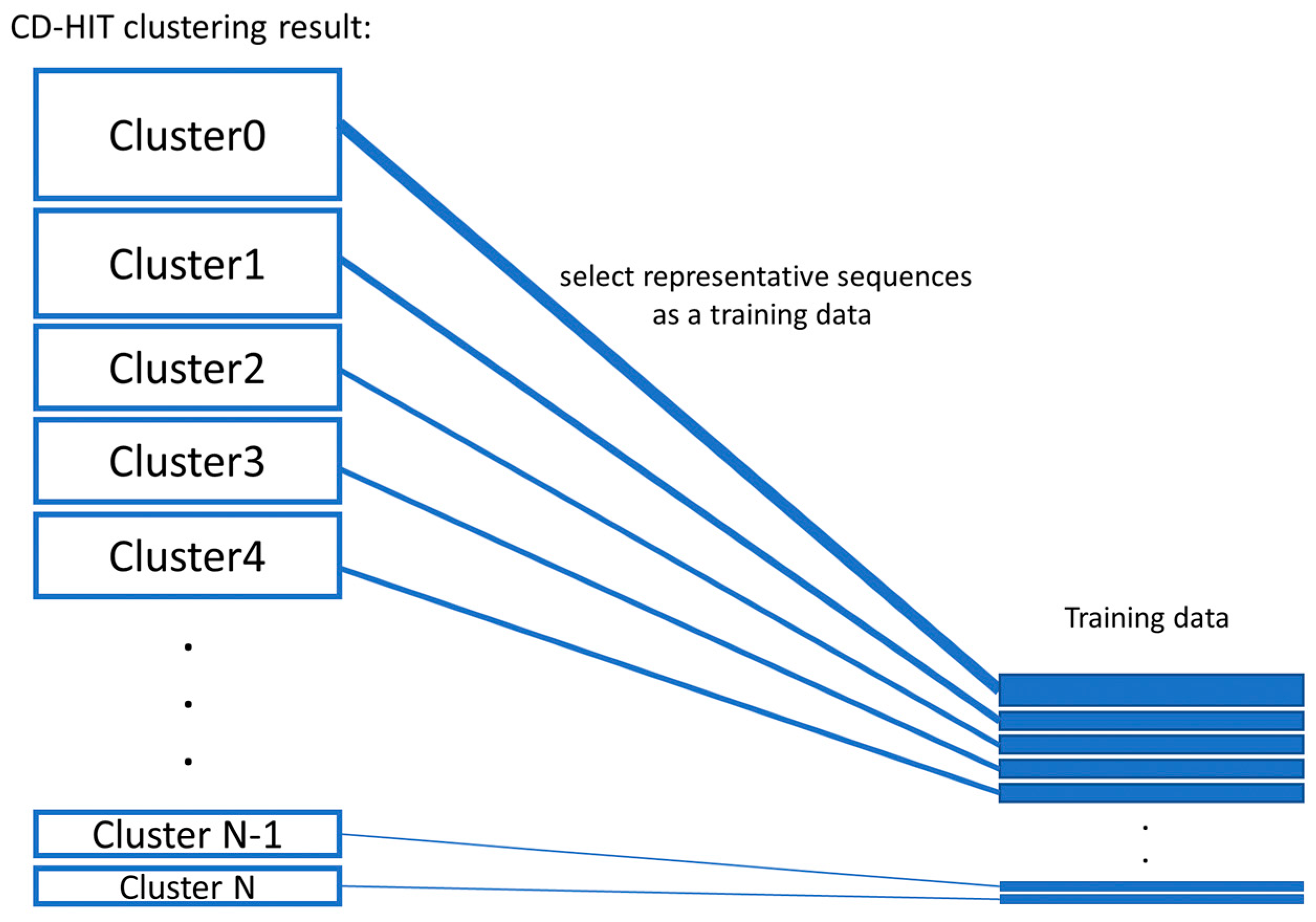





![Swarm: robust and fast clustering method for amplicon-based studies [PeerJ] Swarm: robust and fast clustering method for amplicon-based studies [PeerJ]](https://dfzljdn9uc3pi.cloudfront.net/2014/593/1/fig-2-full.png)